Supervisor: Dr Xavier Bisteau
Esophageal squamous cell carcinoma (eSCC) remains among the deadliest cancers worldwide due to its high mutation burden and heterogeneous landscape of oncogenic alterations. Many reports for this cancer over the years have depicted promising therapeutic options using in vitro 2D culture system but often failed to be translated or even confirmed in clinics because of their lack of physiological complexity similar to tumors. Developing a suitable in vivo models similar and applicable to tumor types remains a challenge. Inversely, in vitro 3D tumor models of human cancer, especially for eSCC are significant tools for investigating cell behavior in a multicellular environment, which can closely mimic the biological and physiological reality. This next generation of in vitro systems recapitulates cell-cell interactions and the complexity found in tumors in more details compared to 2D cultures, offering the possibility of a deeper study of tumor progression and enables drug screening without the involvement of animal models, in particular in the early stages of screening.
Our group has recently developed an in vitro system mimicking a vascularized tumoroid of eSCC where we can observe the interaction of multiple cell types (cancer cells, fibroblast, endothelial cells and immune cells). We intend to understand the interaction of the different cell types, their fate and their molecular behavior upon different treatments. The work in our lab has pointed out that the inhibition of CDK4/6 kinases are promising in vitro using 2D cultures. More importantly, we evidenced that the treated cancer cells induced immunogenic signals. Using this novel in vitro 3D model, we want to understand the causes and the consequences of this immunogenic signals by evaluating the proteome, phosphoproteome and kinome of the different cell types using mass spectrometry.
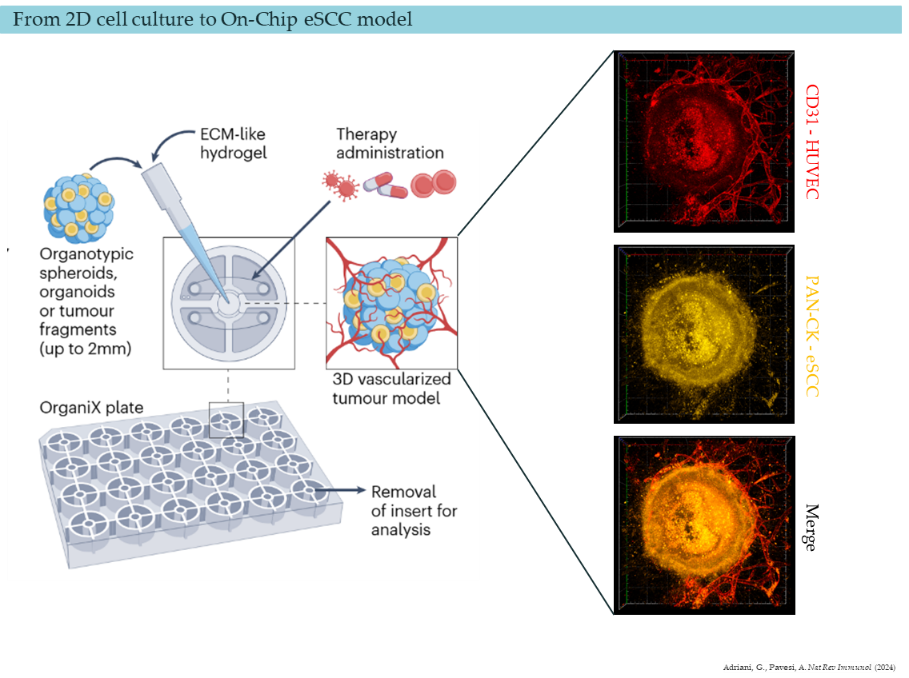
As a PhD candidate joining our dynamic research team, you will have the opportunity to delve into cutting-edge research focusing on three-dimensional culture models of esophageal epithelium and tumors. Your primary objectives will include studying the response of cancer cells, immune cells and fibroblasts to different kinase inhibitors, and evaluating the underlying mechanisms at proteomic level driving the cell interactions.
Techniques:
- Multicellular 3D tumoroids cultures
- Proteomics, phosphoproteomics and kinomics by mass spectrometry
- Bioinformatics analysis
- Transcriptomics RNA-seq, genomic analysis
- Western blotting, qPCR, immunofluorescence, flow cytometry, …